New and Advanced Technology in Plant Breeding: How They Can Boost Crop Production and Resilience
Plant breeding is the science and art of improving crop varieties for human use. It has been practiced for thousands of years by farmers and gardeners, who selected and propagated plants with desirable traits such as yield, quality, disease resistance, and adaptation to local conditions. However, traditional plant breeding methods are often slow, labor-intensive, and limited by the available genetic diversity within a species.
In recent decades, new technologies have emerged that can accelerate plant breeding and expand the genetic potential of crops. These technologies include genomic-assisted breeding (GAB), genome editing, speed breeding, high-throughput phenotyping, and artificial intelligence (AI). These technologies can help breeders to create novel crop varieties that can cope with the challenges of climate change, pests, diseases, and food security.
Genomic-Assisted Breeding (GAB)
GAB is the use of molecular markers and genomic information to select plants with desirable traits. Molecular markers are DNA sequences that are associated with specific genes or traits. By screening plants for these markers, breeders can identify and select plants that carry the desired genes or traits without having to wait for them to express in the field. This can save time, resources, and increase the accuracy and efficiency of breeding.
GAB also allows breeders to access the full landscape of genetic diversity within a species by constructing pan-genomes. Pan-genomes are collections of all the genes and genetic variations found in different individuals or populations of a species. By comparing pan-genomes, breeders can identify rare or lost genes or variations that can be used to improve crop performance or introduce new traits.
Genome Editing
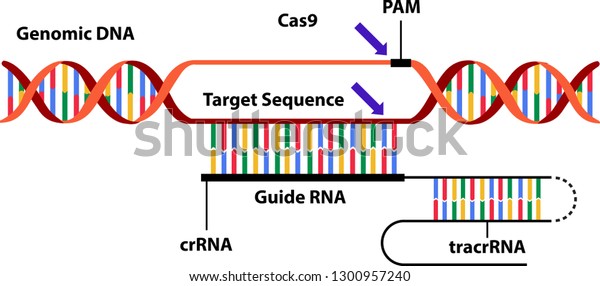
Genome editing is the precise modification of DNA sequences in living cells using engineered nucleases or enzymes that can cut and paste DNA. Genome editing can be used to introduce, delete, or replace specific genes or DNA segments in plants. This can create new variations or traits that are not possible or difficult to achieve by conventional breeding or genetic modification.
One of the most widely used genome editing tools is CRISPR/Cas, which stands for clustered regularly interspaced short palindromic repeats/CRISPR-associated. CRISPR/Cas is a system that consists of a guide RNA that recognizes a target DNA sequence and a Cas enzyme that cuts the DNA at that site. By providing different guide RNAs and Cas enzymes, breeders can edit multiple genes or sites in a plant genome.
Genome editing has numerous applications in crop improvement, such as creating resistance to abiotic and biotic stress, enhancing nutritional quality, modifying plant architecture, and facilitating domestication. Genome editing can also be combined with other techniques such as base editing, prime editing, Cisgenesis, intragenesis, oligonucleotide-directed mutagenesis, reverse breeding, and agro-infiltration to create more precise and diverse changes in plant genomes.
Speed Breeding
Speed breeding is the use of controlled environmental conditions such as light, temperature, humidity, and nutrition to shorten the life cycle of plants and increase the number of generations per year. Speed breeding can accelerate the development and evaluation of new crop varieties by reducing the time required for flowering, seed production, and seed germination.
Speed breeding can be applied to various crops such as cereals, legumes, oilseeds, vegetables, and ornamentals. Speed breeding can also be integrated with GAB and genome editing to rapidly introduce and test new traits in plants. Speed breeding can also enable breeders to exploit the natural variations in underutilized crops that have potential for adaptation to changing climates.
High-Throughput Phenotyping
High-throughput phenotyping is the use of automated or semi-automated methods to measure plant traits such as growth, morphology, physiology, biochemistry, and yield. High-throughput phenotyping can generate large amounts of data on plant performance under different environmental conditions. This data can help breeders to identify and select plants with superior traits or stress tolerance.
High-throughput phenotyping can be performed at different scales such as laboratory, greenhouse, field, or aerial platforms. High-throughput phenotyping can also employ various sensors or imaging techniques such as cameras, spectrometers, lidars, radars, thermometers, fluorometers, chlorophyll meters, gas analyzers, etc. High-throughput phenotyping can also be coupled with AI and machine learning to analyze and interpret the data and provide insights for breeding decisions.
Artificial Intelligence (AI)
Artificial intelligence refers to the simulation of human intelligence processes by machines. In plant breeding, AI can optimize breeding programs by utilizing tools such as data mining, pattern recognition, prediction modeling, optimization algorithms, simulation modeling, and decision support systems. AI helps breeders integrate and analyze large and complex datasets from genomics, Phenomics, environment, and management. It also aids in designing and executing more efficient experiments and trials. AI can discover new genes, traits, or interactions that enhance crop performance and resilience.
AI can help breeders to integrate and analyze large and complex data sets from various sources such as genomics, Phenomics, environment, and management. AI can also help breeders to design and execute more efficient and effective experiments and trials. AI can also help breeders to discover new genes, traits, or interactions that can improve crop performance or resilience.
Conclusion
New and advanced technologies in plant breeding can offer great opportunities for creating novel and improved crop varieties that can meet the current and future demands of agriculture and food security. These technologies can also help breeders to overcome the limitations and challenges of traditional plant breeding methods. However, these technologies also pose some challenges such as ethical, social, legal, regulatory, and economic issues that need to be addressed by stakeholders and policymakers. Therefore, it is important to foster a constructive dialogue and collaboration among researchers, breeders, farmers, consumers, regulators, and society to ensure the safe and responsible use of these technologies for the benefit of humanity and the environment.
Some examples of crops that have been improved using these technologies are:
Corn: Bayer has used precision breeding and artificial intelligence to create corn varieties that are tailored to specific field conditions and customer needs. Bayer has also used genome editing to create corn varieties that are resistant to drought and herbicides.
Wheat: Researchers have used speed breeding and genome editing to create wheat varieties that are resistant to diseases, pests, and heat stress. They have also used genome editing to modify the gluten content and quality of wheat.
Rice: Researchers have used genomic-assisted breeding and high-throughput phenotyping to create rice varieties that are tolerant to salinity, drought, flooding, and cold stress. They have also used genome editing to create rice varieties that are resistant to bacterial blight and have enhanced nutritional value⁴⁵.
Tomato: Researchers have used genome editing and morphogenic factors to create tomato varieties that have improved fruit size, shape, color, flavor, and shelf life. They have also used genome editing to create tomato varieties that are resistant to viruses and nematodes .
Potato: Researchers have used Cisgenesis and intragenesis to create potato varieties that are resistant to late blight, a devastating fungal disease. They have also used oligonucleotide-directed mutagenesis to create potato varieties that have reduced acrylamide content, a potential carcinogen .
Source:
(1) New Technologies Driving the Future of Plant Breeding - Bayer. https://www.bayer.com/en/agriculture/new-technologies-driving-future-plant-breeding.
(2) Recent advances in crop transformation technologies - Nature. https://www.nature.com/articles/s41477-022-01295-8.
(3) Advances in Crop Breeding Through Precision Genome Editing. https://www.frontiersin.org/articles/10.3389/fgene.2022.880195/full.
(4) (PDF) Biotechnology: An Advanced Tool for Crop Improvement - ResearchGate. https://www.researchgate.net/publication/331540922_Biotechnology_An_Advanced_Tool_for_Crop_Improvement.
(5) Next-Generation Breeding Strategies for Climate-Ready Crops. https://www.frontiersin.org/articles/10.3389/fpls.2021.620420/full.
(1) New Plant-Breeding Techniques: What are we talking about?. https://www.farm-europe.eu/travaux/new-plant-breeding-techniques-what-are-we-talking-about/.
(2) New Technologies Driving the Future of Plant Breeding - Bayer. https://www.bayer.com/en/agriculture/new-technologies-driving-future-plant-breeding.
(3) Next-Generation Breeding Strategies for Climate-Ready Crops. https://www.frontiersin.org/articles/10.3389/fpls.2021.620420/full.
(4) New plant breeding techniques and their regulatory ... - PubMed. https://pubmed.ncbi.nlm.nih.gov/33631493/.
(5) Accelerated Breeding of Plants: Methods and Applications. https://link.springer.com/chapter/10.1007/978-3-030-41866-3_1.
(6) Role of New Plant Breeding Technologies for Food Security and .... https://onlinelibrary.wiley.com/doi/10.1002/aepp.13044.